Why are there still over 1,000 uncharacterized yeast genes? |
|
| Lourdes
Peña-Castillo and Timothy R. Hughes* Banting and Best Department of Medical Research University of Toronto 160 College St., Toronto, ON M5S3E1, Canada |
|
| *To whom
correspondence should be addressed: t.hughes at utoronto.ca TEL: 416-946-8260 FAX: 416-978-8528 |
|
 |
|
Abstract |
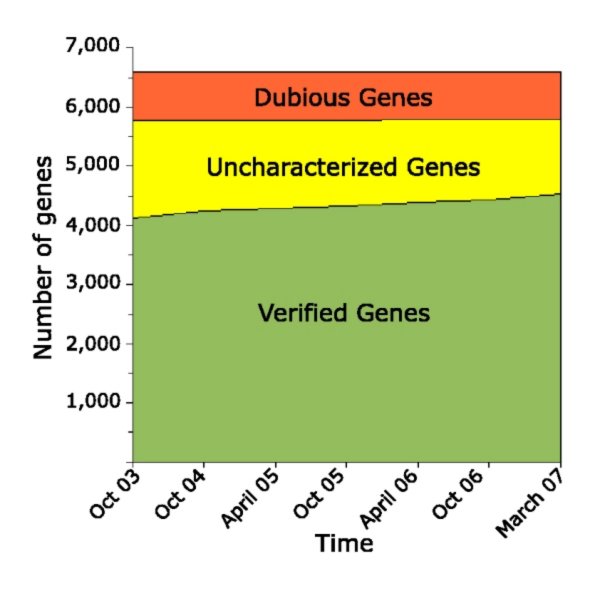
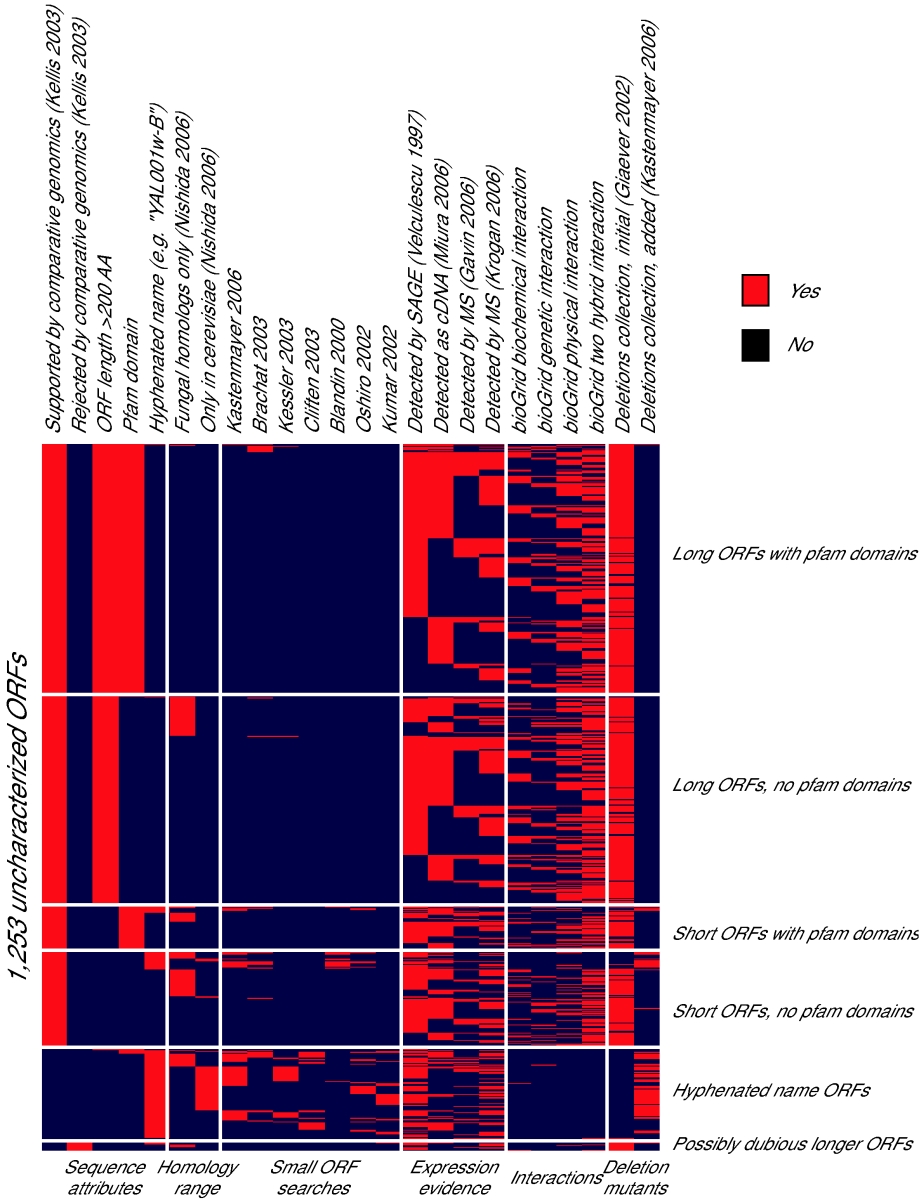
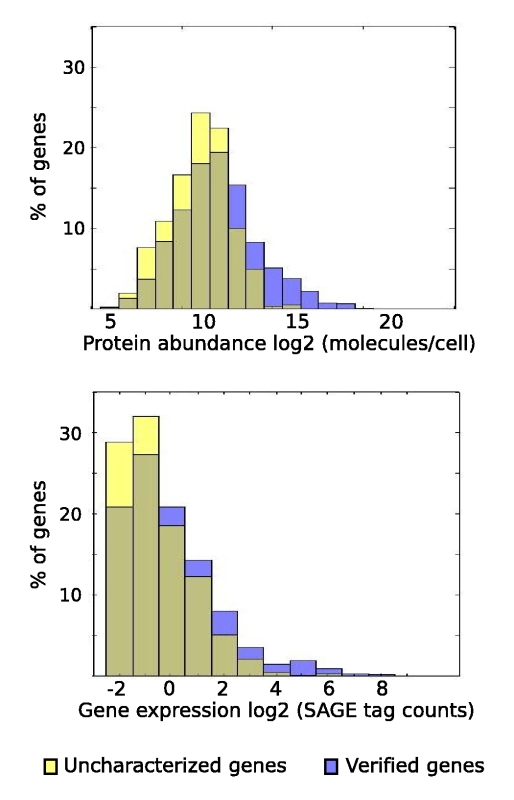
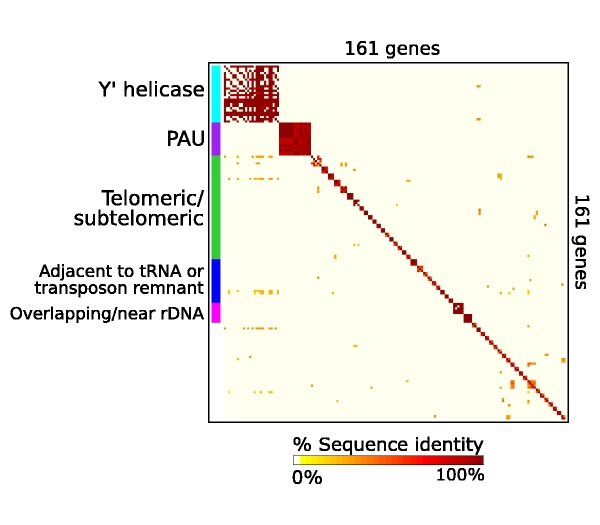
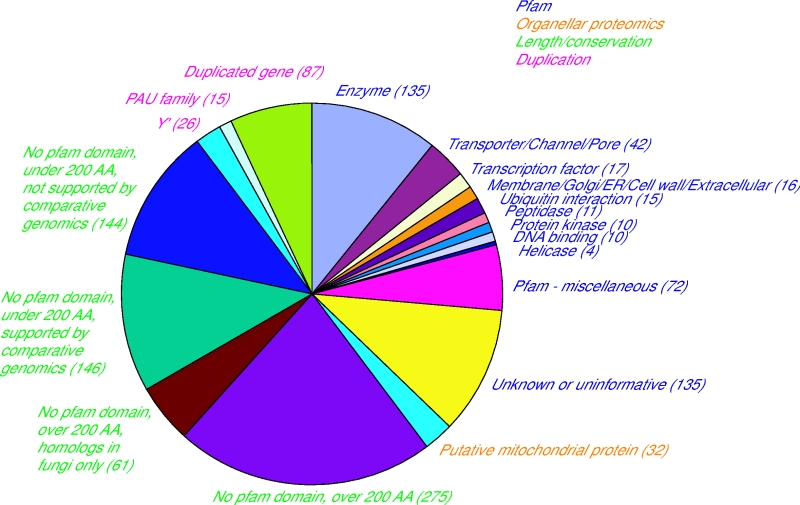
The yeast genetics community has embraced genomic biology, and there is a general understanding that obtaining a full encyclopedia of functions of the ~6,000 genes is a worthwhile goal. The yeast literature comprises over 40,000 research papers, and the number of yeast researchers exceeds the number of genes. There are mutated and tagged alleles for virtually every gene, and hundreds of high-throughput data sets and computational analyses have been described. Why, then, are there over 1,000 genes still listed as uncharacterized on Saccharomyces Genome Database, ten years after sequencing the genome of this powerful model organism? Examination of the currently-uncharacterized gene set suggests that while some are small or newly-discovered, the vast majority were evident from the initial genome sequence. Most are present in multiple genomics data sets, which may provide clues to function. In addition, roughly half contain recognizable protein domains, and many of these suggest specific metabolic activities. Notably, the uncharacterized gene set is highly enriched for genes whose only homologues are in other fungi. Achieving a full catalogue of yeast gene functions may require a greater focus on the life of yeast outside the laboratory. |
|
|
|
List of uncharacterized genes in SGD as of March 20, 2007 |
|
| Number
of interactions, papers, GO annotations per gene |
|
|
|
|
|
EPS Data in Excel file |
 |
|
EPS Data in Excel file |
 |
|
EPS Data in Excel file |
 |
|
EPS Data in Excel file |
 |
|
EPS Data in Excel file |
 |
Last updated on Tue 22 May, 2007