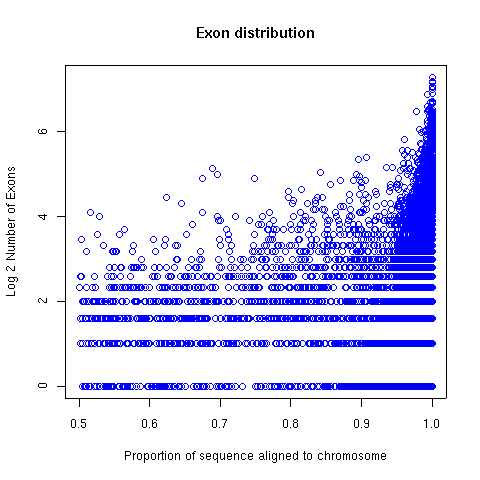
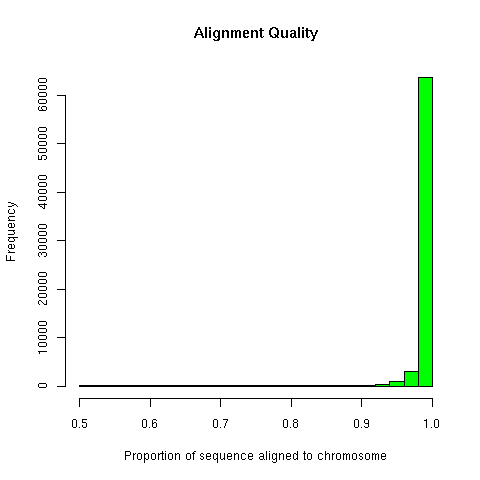
Exon foud by aligning to the mouse genome (NCBI build
36, Feb.
2006), with a mininum allowed exon length of 20nt and a minimum allowed
intron size of 50nt, the following:
- 48,360
unique IRC sequences. From those, 47,927 IRC
sequences whose best alignment with a chromosome involves at least 50%
of the IRC sequence were taken for exon-tiling.
- 10,471 RefSeq sequences not contained in the exon
collection obtained in step 1 (according to blat alignments). From those, 10,095 RefSeq sequences
whose best alignment with a chromosome involves at least 50% of the
RefSeq
sequence were taken for exon-tiling.
- 11,257 Ensembl transcripts (9,831 Ensembl genes) not
contained in the exon collection obtained in steps 1 and 2 (according to MGI-Ensembl tables provided by MGI). From those,
11,110 transcripts whose best alignment with a chromosome involves at
least 50% of the transcript sequence were taken for exon-tiling.
| Source |
Number of
sequences taken for exon-tiling |
Number of
Exons obtained |
Median Number of Exons per sequence |
Mean Number of Exons per Sequence |
Maximum Number of Exons per Sequence |
| IRC |
47,927 |
353,809 |
5 |
7.38 |
155 |
| RefSeq |
10,095 |
25,199 |
2 |
2.49 |
79 |
| Ensembl |
11,110 |
54,365 |
2 |
4.89 |
147 |
| Total |
69,132 |
433,373 |
4 |
6.27 |
155 |
|